Bridges
This picture is almost 11 years old. Too old, if you ask me.
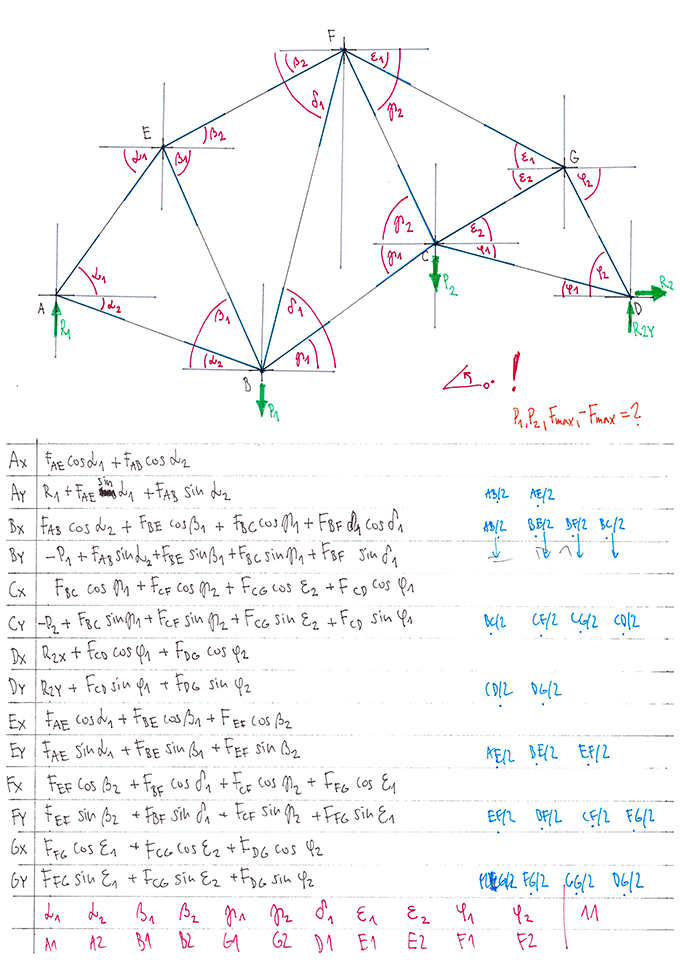
During the winter 2002/2003 I was working on a school project regarding the topic "bridges". Good "bridges" are better than bad ones so I set out to find an optimal configuration of a truss bridge. The basic principle was to minimize the compression/tension force in the most stressed member, i.e. to distribute the load among different members. One way of doing it, and for which the source code is displayed below, consists of two parts: calculating the static scheme of a structure and exploring the search space with genetical algorithm.
To say that a structure is stable is to say that the forces acting on all the joints are in static equilibrium — they all zero out. Thus one can write down the problem as a system of linear equations and solve it with Gaussian elimination. As truss bridges fail when one of its members fails, the smaller is the force acting upon the most stress member, the better. The question then is, where to put the joints so that the load spreads out nicely?
Genetical algorithm (with elitism) is a stochastic method for locating the global extrema of a function. While applying the natural selection pressure, it cross-breeds and mutates a population of states. The better an individual fits in, the greater chance it has to mate with another fitting individual, hence to carry part of its genome to next generation. In the example of bridges, the fitness criterion is then the inverse of most intense force. To stop the evolutionary process one can look for the magnitude of improvement along the generations. Or to mind the width of the poster.
Animations capturing the bridge optimization (four runs):
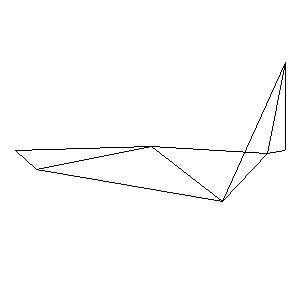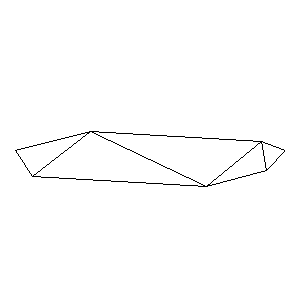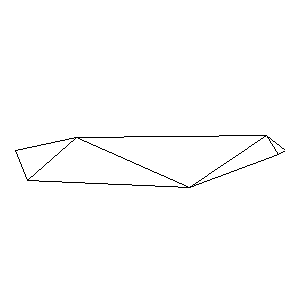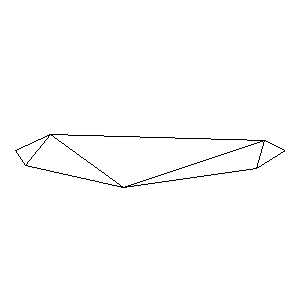
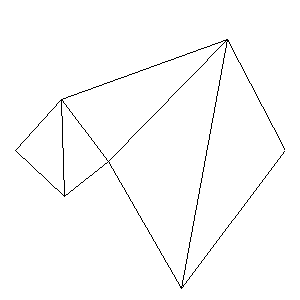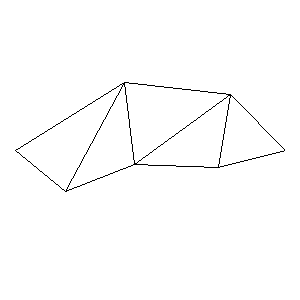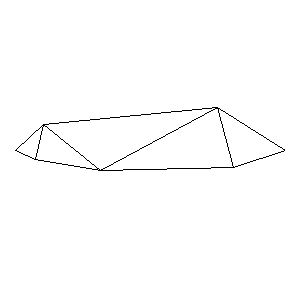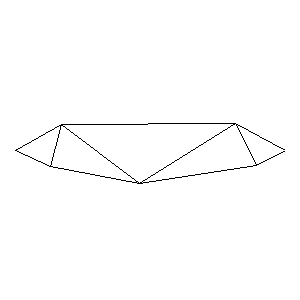
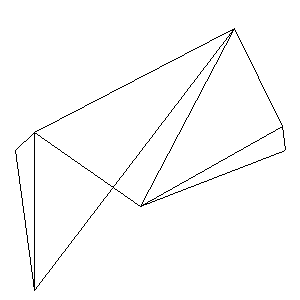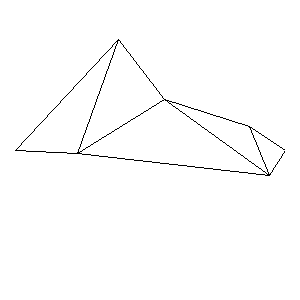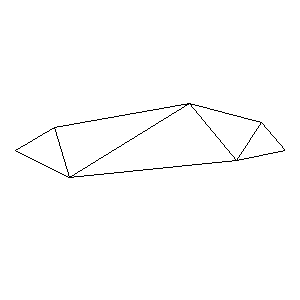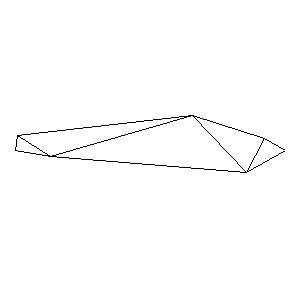
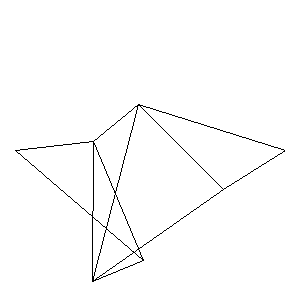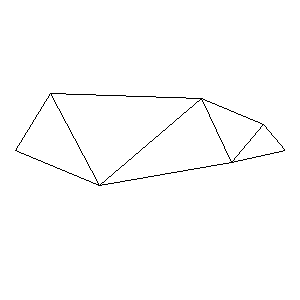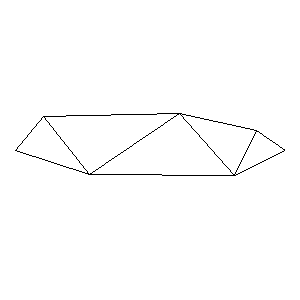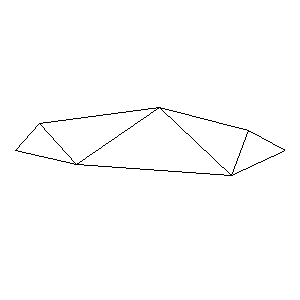
Original source code (for Python 2.2!) and cleaned-up version using Flat:
from bisect import bisect_left
from math import hypot
from random import normalvariate, random, randrange
from flat import shape, document, view
def solve(a, b):
# Gaussian elimination with partial pivoting
n = len(a)
assert n == len(a[0]), 'Non-square matrix.'
assert n == len(b), 'Invalid dimensions.'
for k in range(n - 1):
pivot, value = k, abs(a[k][k])
for j in range(k + 1, n):
v = abs(a[j][k])
if v > value:
pivot, value = j, v
if pivot != k:
a[pivot], a[k] = a[k], a[pivot]
b[pivot], b[k] = b[k], b[pivot]
for j in range(k + 1, n):
scale = a[j][k] / a[k][k]
for i in range(k + 1, n):
a[j][i] -= a[k][i] * scale
b[j] -= b[k] * scale
x = [0.0] * n
for j in range(n - 1, -1, -1):
for i in range(j + 1, n):
b[j] -= a[j][i] * x[i]
x[j] = b[j] / a[j][j]
return x
def members(genomes, x):
for y, (ax,ay,ex,ey,bx,by,fx,fy,cx,cy,gx,gy,dx,dy) in enumerate(genomes):
yield ax+x, ay+y, ex+x, ey+y
yield ax+x, ay+y, bx+x, by+y
yield ex+x, ey+y, bx+x, by+y
yield ex+x, ey+y, fx+x, fy+y
yield bx+x, by+y, fx+x, fy+y
yield bx+x, by+y, cx+x, cy+y
yield fx+x, fy+y, cx+x, cy+y
yield fx+x, fy+y, gx+x, gy+y
yield cx+x, cy+y, gx+x, gy+y
yield cx+x, cy+y, dx+x, dy+y
yield gx+x, gy+y, dx+x, dy+y
def forces(genomes):
for ax, ay, ex, ey, bx, by, fx, fy, cx, cy, gx, gy, dx, dy in genomes:
ab = hypot(ax-bx, ay-by)
ae = hypot(ax-ex, ay-ey)
bc = hypot(bx-cx, by-cy)
be = hypot(bx-ex, by-ey)
bf = hypot(bx-fx, by-fy)
cd = hypot(cx-dx, cy-dy)
cf = hypot(cx-fx, cy-fy)
cg = hypot(cx-gx, cy-gy)
dg = hypot(dx-gx, dy-gy)
ef = hypot(ex-fx, ey-fy)
fg = hypot(fx-gx, fy-gy)
ca1a, sa1a = (ex-ax)/ae, (ey-ay)/ae
ca1e, sa1e = (ax-ex)/ae, (ay-ey)/ae
ca2a, sa2a = (bx-ax)/ab, (by-ay)/ab
ca2b, sa2b = (ax-bx)/ab, (ay-by)/ab
cb1b, sb1b = (ex-bx)/be, (ey-by)/be
cb1e, sb1e = (bx-ex)/be, (by-ey)/be
cb2e, sb2e = (fx-ex)/ef, (fy-ey)/ef
cb2f, sb2f = (ex-fx)/ef, (ey-fy)/ef
cg1b, sg1b = (cx-bx)/bc, (cy-by)/bc
cg1c, sg1c = (bx-cx)/bc, (by-cy)/bc
cg2c, sg2c = (fx-cx)/cf, (fy-cy)/cf
cg2f, sg2f = (cx-fx)/cf, (cy-fy)/cf
cd1b, sd1b = (fx-bx)/bf, (fy-by)/bf
cd1f, sd1f = (bx-fx)/bf, (by-fy)/bf
ce1f, se1f = (gx-fx)/fg, (gy-fy)/fg
ce1g, se1g = (fx-gx)/fg, (fy-gy)/fg
ce2c, se2c = (gx-cx)/cg, (gy-cy)/cg
ce2g, se2g = (cx-gx)/cg, (cy-gy)/cg
cf1c, sf1c = (dx-cx)/cd, (dy-cy)/cd
cf1d, sf1d = (cx-dx)/cd, (cy-dy)/cd
cf2d, sf2d = (gx-dx)/dg, (gy-dy)/dg
cf2g, sf2g = (dx-gx)/dg, (dy-gy)/dg
p1, p2, f = 10.0, 10.0, 1000.0
a = [
[ca2a,ca1a, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],
[sa2a,sa1a, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0],
[ca2b, 0.0,cg1b,cb1b,cd1b, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],
[sa2b, 0.0,sg1b,sb1b,sd1b, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],
[ 0.0, 0.0,cg1c, 0.0, 0.0,cf1c,cg2c,ce2c, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],
[ 0.0, 0.0,sg1c, 0.0, 0.0,sf1c,sg2c,se2c, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],
[ 0.0, 0.0, 0.0, 0.0, 0.0,cf1d, 0.0, 0.0,cf2d, 0.0, 0.0, 0.0, 1.0, 0.0],
[ 0.0, 0.0, 0.0, 0.0, 0.0,sf1d, 0.0, 0.0,sf2d, 0.0, 0.0, 0.0, 0.0, 1.0],
[ 0.0,ca1e, 0.0,cb1e, 0.0, 0.0, 0.0, 0.0, 0.0,cb2e, 0.0, 0.0, 0.0, 0.0],
[ 0.0,sa1e, 0.0,sb1e, 0.0, 0.0, 0.0, 0.0, 0.0,sb2e, 0.0, 0.0, 0.0, 0.0],
[ 0.0, 0.0, 0.0, 0.0,cd1f, 0.0,cg2f, 0.0, 0.0,cb2f,ce1f, 0.0, 0.0, 0.0],
[ 0.0, 0.0, 0.0, 0.0,sd1f, 0.0,sg2f, 0.0, 0.0,sb2f,se1f, 0.0, 0.0, 0.0],
[ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,ce2g,cf2g, 0.0,ce1g, 0.0, 0.0, 0.0],
[ 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,se2g,sf2g, 0.0,se1g, 0.0, 0.0, 0.0]]
b = [
0.0, ab*f + ae*f,
0.0, p1 + ab*f + be*f + bf*f + bc*f,
0.0, p2 + bc*f + cf*f + cg*f + cd*f,
0.0, cd*f + dg*f,
0.0, ae*f + be*f + ef*f,
0.0, ef*f + bf*f + cf*f + fg*f,
0.0, fg*f + cg*f + dg*f]
x = solve(a, b)
yield max(abs(max(x)), abs(min(x)))
population, generations = 100, 60
figure = shape().width(0.2)
doc = document(generations, population, 'cm')
page = doc.addpage()
genomes = [[
0.05, 0.5,
random(), random(),
random(), random(),
random(), random(),
random(), random(),
random(), random(),
0.95, 0.5] for i in range(population)]
for x0, y0, x1, y1 in members(genomes, 0):
page.place(figure.line(x0, y0, x1, y1))
for generation in range(1, generations):
fitness = [1.0 / f ** 2 for f in forces(genomes)]
best = fitness.index(max(fitness))
for i in range(1, population):
fitness[i] += fitness[i - 1]
new = [genomes[best]]
for i in range(1, population):
a = bisect_left(fitness, random() * fitness[-1])
b = bisect_left(fitness, random() * fitness[-1])
split = randrange(0, 15)
new.append(genomes[a][:split] + genomes[b][split:])
genomes = new
for i in range(population):
u, v = randrange(1, population), randrange(2, 12)
genomes[u][v] = normalvariate(genomes[u][v], 0.1)
if genomes[u][v] < 0.0:
genomes[u][v] = -genomes[u][v]
elif genomes[u][v] > 1.0:
genomes[u][v] = 2.0 - genomes[u][v]
for i in range(5):
u, v = randrange(1, population), randrange(2, 12)
genomes[u][v] = random()
for x0, y0, x1, y1 in members(genomes, generation):
page.place(figure.line(x0, y0, x1, y1))
view(doc.pdf())
— 20. 11. 2013